by nikhil bhatla, nbhatla@mit.edu, nikhil.superfacts.org
June 18, 2009 (updated July 26, 2012, again on Oct 17, 2016)
An Interactive Visualization of the C. elegans Neural Network
(Works in Firefox, Google Chrome, and Safari, but not Internet Explorer)I wrote this page for readers who are C. elegans researchers. If you'd like a more basic introduction to C. elegans, read Visualizing a Worm's Neural Network.
Features:
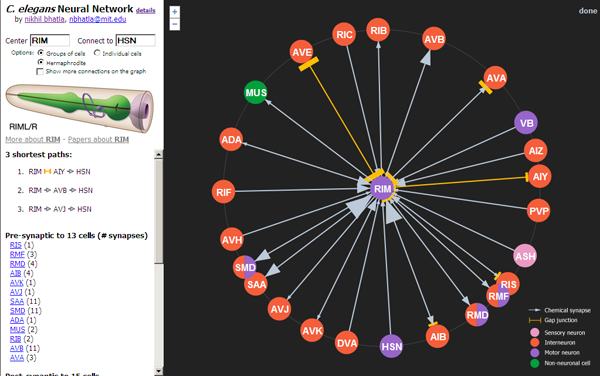
- Click on neurons to re-center them and see what other neurons they're connected to.
- Specify a start neuron and an end neuron to find the shortest path between 2 neurons (e.g. BAG [putative carbon dioxide sensor] and HSN [egg-laying motor neuron]).
- Zoom in and out to view neurons more than just 1-degree away.
- View all connections in the visible portion of the network (makes a dense spiderweb) by checking the "Show more connections on the graph" checkbox.
- View and search the network of individual neurons. By default, neuron groups (e.g. pairs of neurons) are displayed, but this is an oversimplification, and the individual neuron network is a more accurate representation of the true network.
Cells included: (parentheses indicate how to search for the cell type)
- 302 individual neurons in 118 groups
- 1 neuron-like cell class (XXX)
- 7 pharyngeal muscles (PM)
- 2 pharyngeal marginal cells (PMC)
- 2 pharyngeal gland cells (PGC)
- 1 pharyngeal basement membrane (PBM)
- 1 muscle class (MUS) that includes body wall muscles, vulval muscles and anal muscles
- 1 intestine class (INT)
Data sources:
- The main network data is from the Chklovskii group and was published in Chen..Chklovskii 2006. I modified the raw data file a bit to ease the processing of it, and it is available here. Details for how to read the file are here. A list of neurons, non-neuronal interacting cells, and the associated groups are here, modified from the Japanese CCEP group. Note that this data file includes the following primary work:
- White..Brenner 1986.
- 5 animals were reconstructed in this paper: 3 adult hermaphrodites (N2T, N2U, JSE), 1 L4 larva (JSH), and 1 adult male (N2Y). The connectivity data used by the Chklovskii group (and hence used in this visualization) was primarily derived from adults N2U and JSE. Area anterior to the nerve ring was covered with adult N2T, and the gap between N2U and JSE is covered by adult male N2Y. L4 JSH was only used as a check of nerve ring connectivity. So, in actuality, the visualized network is a hermaphrodite-male hybrid and a 5-worm hybrid.
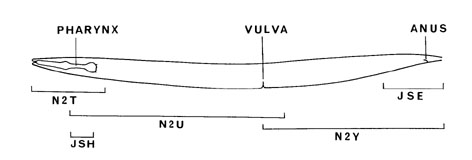
- 5 animals were reconstructed in this paper: 3 adult hermaphrodites (N2T, N2U, JSE), 1 L4 larva (JSH), and 1 adult male (N2Y). The connectivity data used by the Chklovskii group (and hence used in this visualization) was primarily derived from adults N2U and JSE. Area anterior to the nerve ring was covered with adult N2T, and the gap between N2U and JSE is covered by adult male N2Y. L4 JSH was only used as a check of nerve ring connectivity. So, in actuality, the visualized network is a hermaphrodite-male hybrid and a 5-worm hybrid.
- Durbin 1986
- Hall & Russell 1991
- Hobert & Hall 1999
- Achacoso & Yamamoto 1992
- White..Brenner 1986.
- The pharynx network data is from Albertson & Thomson 1976. At least 2 animals were compared for all observations from this paper. My data file is here, which was modified from the Japanese CCEP group's version. This data is older and much less clean than the first data set. Some of the issues are:
- Some synapses are recorded as being between a single cell and a cell group. In these cases, connections are assumed to be to all cells of that group (the most conservative assumption given the data, in my opinion).
- Sometimes the type of synapse is missing. In these cases, I either default to a pre-synapse, or use the comment made in the text (but missing in the figure).
- A synapse is sometimes annotated as both a gap junction and a chemical synapse. In these cases, I assume both are present. This causes double-counting of some synapses, so the synapse count for the pharynx isn't very accurate.
- Often the target muscle name is missing. In these cases, I direct the synapse to the muscle that I infer from the figure.
- Sometimes the annotations from CCEP were wrong, so I fixed them.
- I renamed several of the muscles and other non-neuronal cells to names that don't collide with and are more consistent with main network neurons.
- I count each line of the data file as a single synapse.
- Many synapses in the pharynx are highly variable from animal to animal (i.e. sometimes they're there and sometimes they aren't), as noted in the data file. These are included in the visualization.
Visualization:
- For the fancy animation and radial graph, I built off of an existing javascript library called JIT. I modified only a small part of it to accommodate my use, and greatly appreciate the fact that this library was available, open source, and free to use.
- The drawings of the neurons on the upper left side of the screen come from WormAtlas.
- The size of a grey arrow is proportional to the number of chemical synapses between the 2 cells, and the size of an orange bar is proportional to the number of gap junctions between 2 cells.
Misc. notes:
- The network can be broken into 2 pieces: the main neural network and the phrayngeal neural network. These networks are thought to be connected by only a single gap junction between RIP (of the main network) and I1 (of the pharyngeal network).
- 1 neuron pair (CAN) is not known to make any synapses to other neurons, so is effectively an island in the network.
- The number of animals surveyed for this connectivity chart depends on the part of the worm (see cartoon above). In the nerve ring, 3 animals (2 adults and 1 L4 larva) were studied. Although they don't all share the same synaptic connections (there are substantial differences in synapses and gap junctions made between adult N2U and L4 JSH), they are somewhat consistent. The anal region was studied in 2 adults (JSE and N2Y), and the ventral nerve cord was studied in 2 whole adults (N2U+N2Y and N2S). The region anterior to the nerve ring comes from just 1 adult (N2T). Bottom line: what's shown here should be treated as a sampling, not a definitive network, for all individuals of the Bristol N2 strain of C. elegans.
- Finally, just because 2 neurons are anatomically connected does not mean that they're functionally connected. And just because 2 neurons are NOT anatomically connected does not mean that they are NOT functionally connected.
Known bugs and issues:
- If you're viewing individual neurons (instead of neuron groups), clicking on some neurons in the pharynx can be slow (e.g. MCL). Be patient as it may take several minutes for the network to re-center (but it will eventually!). This may be fixed in the future.
- You can only search for paths between neurons that can be found by the program within 60 seconds, which is a small subset of all possible paths. Sometimes it may timeout, yielding no paths if the neurons are sufficiently far away from each other. This may be fixed in the future.
- The data set isn't 100% accurate. I've recently noticed that some of the Chklovskii data is missing some of the data from the White 1986 paper, data that shouldn't be excluded based on the criteria they provide. For example, the Chklovskii data set is missing a synapse from BAG to AUA which is in White 1986. I may look in to this.
Bug related to synapse counts fixed! (as of Oct 17, 2016)
- Several users reported an issue with the neural network undercounting the number of synapses between two neurons. Specifically, if a polyadic synapse was present from neuron A to neuron B in the Chklovskii dataset, monadic synapses were not included in the synapse count reported by the visualization. By fixing this problem, the total number of synapses in the visualization has increased from 9,237 to 10,204, an increase of 10%. Roughly 15% of synapse pairs were affected by this bug. This issue did not affect whether two neurons were connected, but did affect the synapse count associated with the connection. In some cases the effect was small (AVA -> PVC went from 21 to 28 synapses), and in other cases the effect was large (AVBL -> AVAL went from 1 to 7 synapses, ASG -> AIA went from 5 to 19 synapses). In all cases the synapse count increased. As of Oct 17, 2016, this bug has been resolved as fixed. Apologies to anyone who was adversely affected by this issue.
Possible future improvements:
- Male-specific network
- Neurotransmitter and receptor expression
I hope you find this interactive neural network interesting, thought-provoking, and, if you're a C. elegans researcher, maybe even useful.